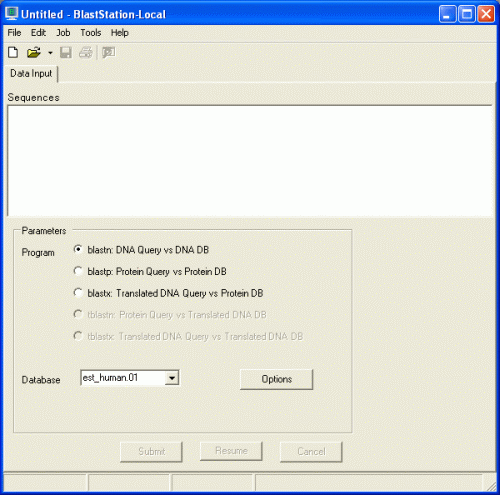
How to start Local BLAST search: Windows
More
detailed help pages are available at
http://www.blaststation.com/help/bslocal/en/win/index.html
or Launch
BlastStation-Local and goto Help > Help Menu.
Double click
BlastStation-Local
icon o the desktop.
 |
|
|
Before performing local BLAST search, you have to download database files from
the NCBI ftp server or create database files from FASTA data.
Downloading Databases
If you always want to perform local BLAST searches against most recent NCBI
databases, Synchronized Database Download function in
BlastStation-Workgroup
is suitable. BlastStation-Workgroup
can perform synchronized download of any FASTA data
on the Internet and create local databases automatically, too.
On the Tools menu, click Download DB. The DB
Download window opens. DB Type column indicates that the database file is in DNA
or Protein database format. For example env_nr.tar.gz is used for blastp and
env_nt.tar.gz is used for blastn.
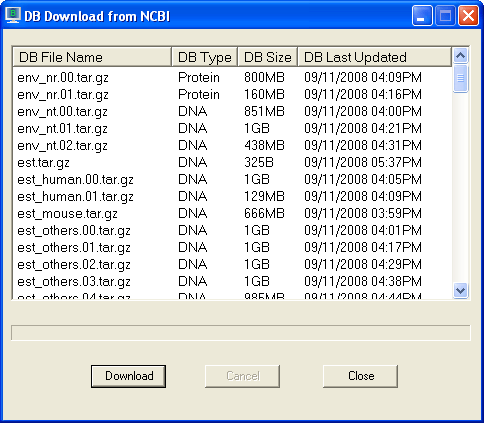
Click database file to highlight and then click "Download" button to download
it. If you want to cancel download, click "Cancel" button during downloading.
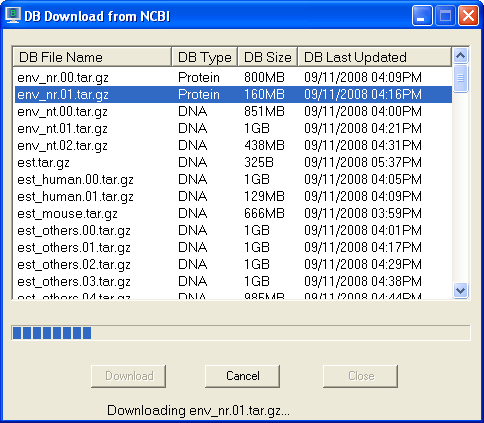
When download is finished, database file will be automatically placed in the
database directory and ready to use.
Creating your own Databases
You can create database files from your
own multi-FASTA data.
FASTA data format is defined here(http://www.ncbi.nlm.nih.gov/blast/html/search.html)
and multi-FASTA data format is defined here(http://baboon.math.berkeley.edu/mavid/multifasta.html).
On the Tools menu, click Create DB. The Create DB window opens.
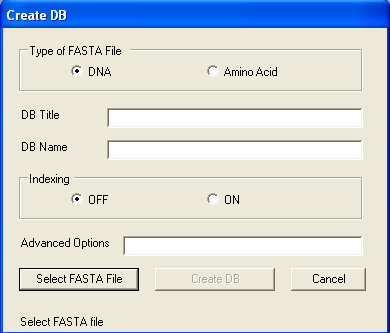
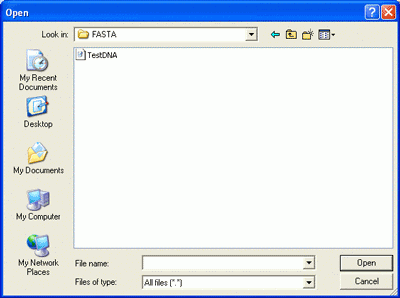
Clicking "Select FASTA File" button will open File Open dialog. Select multi-FASTA
file and click "Open" button.
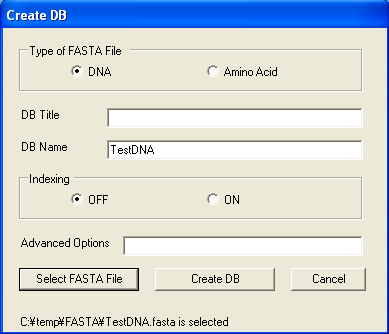
Clicking "Create DB" button creates database. When database is created,
"Database is created" message will be shown. The other options
are for advanced users. Please refer to http://www.blaststation.com/help/bslocal/en/win/Chap2/02_CreateDB.html for details.
BlastStation-Local64 Only
When selected FASTA file is large, multiple database files such as nt.00, nt.01,
and so on will be created. At the same time, the alias file, such as
nt.nal will be created. When this alias file is selected as database,
Blast search over large virtual database can be done.
Start BLAST Search
Copy and paste FASTA data below in the Input
Data.
>gi|11611818|gb|AF287139.1|AF287139 Latimeria chalumnae Hoxa-11 gene,
partial cds
TACTTGCCAAGTTGCACCTACTACGTTTCGGGTCCCGATTTCTCCAGCCTCCCTTCTTTTTTGCCCCAGA
CCCCGTCTTCTCGCCCCATGACATACTCCTATTCGTCTAATCTACCCCAAGTTCAACCTGTGAGAGAAGT
TACCTTCAGGGACTATGCCATTGATACATCCAATAAATGGCATCCCAGAAGCAATTTACCCCATTGCTAC
TCAACAGAGGAGATTCTGCACAGGGACTGCCTAGCAACCACCACCGCTTCAAGCATAGGAGAAATCTTTG
GGAAAGGCAACGCTAACGTCTACCATCCTGGCTCCAGCACCTCTTCTAATTTCTATAACACAGTGGGTAG
AAACGGGGTCCTACCGCAAGCCTTTGACCAGTTTTTCGAGACGGCTTATGGCACAACAGAAAACCACTCT
TCTGACTACTCTGCAGACAAGAATTCCGACAAAATACCTTCGGCAGCAACTTCAAGGTCGGAGACTTGCA
GGGAGACAGACGAGAAGGAGAGACGGGAAGAAAGCAGTAGCCCAGAGTCTTCTTCCGGCAACAATGAGGA
GAAATCAAGCAGTTCCAGTGGTCAACGTACAAGGAAGAAGAGGTGC
>gi|40789109|gb|AC147788.1| Latimeria menadoensis clone VMRC4-19C10,
complete sequence
GAATTCTCGGTCGCTTCTGATCTTCAGTGAAAACTATAAAAGAAAATATAAAAGCAAGCAAGCTACAGAG
TCAGCATAATGCTATAAAACGTAAGGTGAATGAGAAGCAAATGGAAGACAACTAAGAAGATAAACTGTTA
AAGGCCATAAAGTGTACTCGGATAATTTATATATATATATATATTAGAATCACTGAAGAAAACAGCACCT
AGGTGTGTGTTTCGAAAGATGAAATCTACATTTTACGATATTGCTTTCTCCAATGTAAAAGAAGAAACGT
AAATTCCAACAGTATTTAACGCAGAATATATTTTTTCTAACGGTAACACATTAATTTTTTTTTACCCTTT
TCTGGTAGCTTTTTTATTGTTTTAAATTACTACAATCACAATACATTTTAGAGTGATATAAATATGTAAA
ACTAATATACTGTATATATTACTCGTATTGTGTTCTTAGGTCTCAGTATTTTACCTGTGGTGGTGACTGC
TTATCTGTTCAAGGTCACAGAGATATTTATTAAATCATGTTATTTTCGGTTGACTCAGTAAAAGTATAGT
TTGAGTAACAAAAAAGTATCGTGGTTTTGAGATCCTTACTTGGGAAAGGAAGCTAATACATAATTTGCTA
AACTGAGTTTAGGGACAGGTTTATAACTAAAAATGTGAACTACTTCAAATTACCATCATCTACCACATTG
AACGTATTCTTATTATGCATTGAAAATCAATGAATTCGATTACATAAATATACCATAGACTGTTGCATGA
GCAACAGTGGTTCATAGTATTTCTTATTTCTTCATTCAATTGAAGTTTTTGTCATACAGTTTTATTCTAA
CCAAATGATTAATTACTATATATGTGCTGAAAATATAAACATGATTTTCAGTGTATAGGTTGTTAGACCT
CCAGAATGAAAACACAGTTTAGAAACTTGTTTACCTTGCATACAATGTATGTCATTTTTTCTGTATAATT
AAGTTTTGGCATTTGACTTAATGTAATAAATAAGTCGTATATTTAATAATTAAATAATTTCTAATAATCT
GTACTGCTTTTCGATATCTACGTTTTTGAGTAAACTGTTTTCCTCATAAATTCAAAACAAATAAAAAAAG
AGAAATGAAAAAAAAATATTTTCATAACTTGTACTCATTTGCTGAACCAGGTCTTCCCAGGGCTCTTTCG
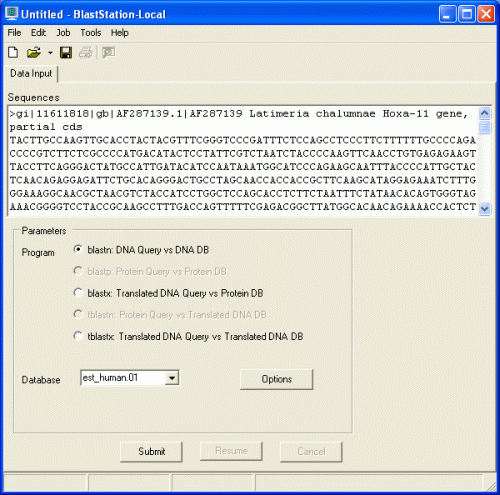
blastn, blastx, and tblastx are available in the Program section.
This is
because the FASTA data are DNA sequences. If the data are protein sequences,
blastp and tblastn will be available.
If blastn is selected, DNA databases
will be shown in the Database pull down menu. If blastp is selected, protein
databases will be shown on the other hand. Two databases, NucleotideExample and
ProteinExample, are installed with BlastStation-Local. You can use them for
testing.
Just click blue "submit" button. The following dialog will
be shown.
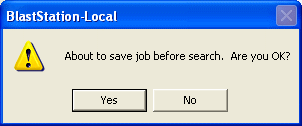
Click "Yes".
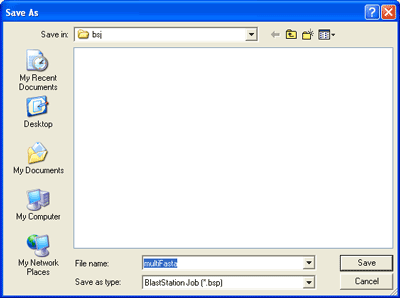
Enter file name and click "Save". This file name will be the BLAST search job
name.
That's it! Now BLAST search is started. The status bar is
showing "Searching...".
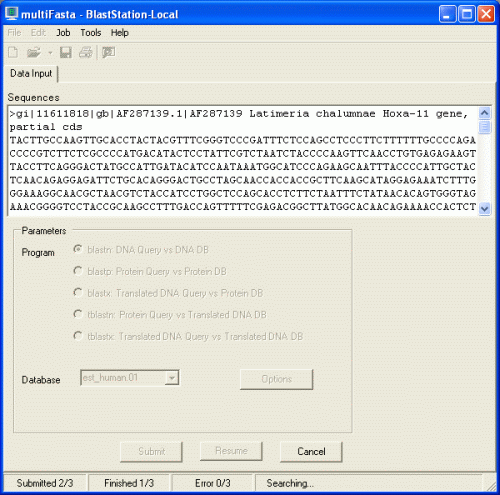
When BLAST search is finished, the following dialog will be displayed.
Click "Yes" to see search results.
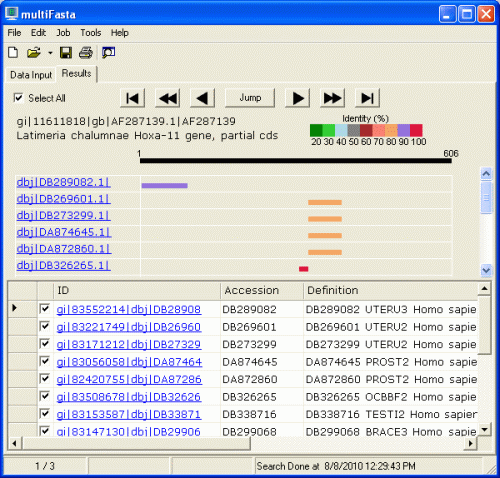
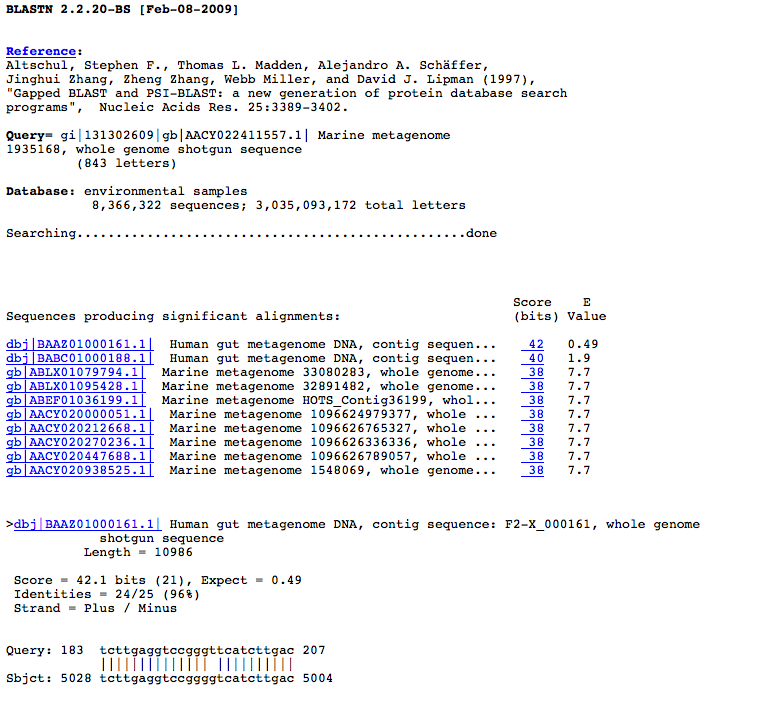
Bar graph is shown on the upper half. The color of the bar is classified by the
identity. Red is 100%, purple is 90% or more, and so on. The length of the query
sequence (606 in this case) is given above the black bar at the top. You can
easily understand which portion of the subject sequence matches with the query
sequence. Move your mouse cursor on the bars. The color of the bar changes and
four numbers will be displayed.
Table is shown on the lower half,
which consists of ID, Accession number, Definition, length, bit score, E Value,
and Identity.
In order to display the other search results, click
arrows below.
 Previous Result
Previous Result
 Next Result
Next Result
 Fast Backward
Fast Backward
 Fast Forward
Fast Forward
 First Result
First Result
 Last Result
Last Result
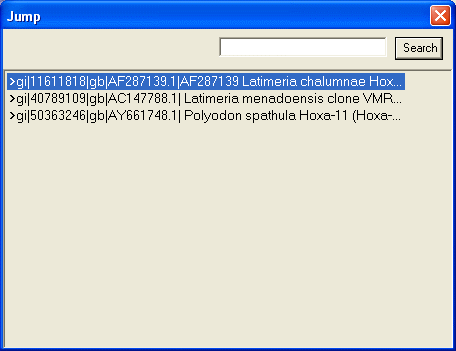
When you click "Jump", the window below will be shown.
Description of each FASTA file will be listed in this window. Clicking each
description will display corresponding search result. If you have many
descriptions, type in search strings in the text field and click search button
to narrow this list.
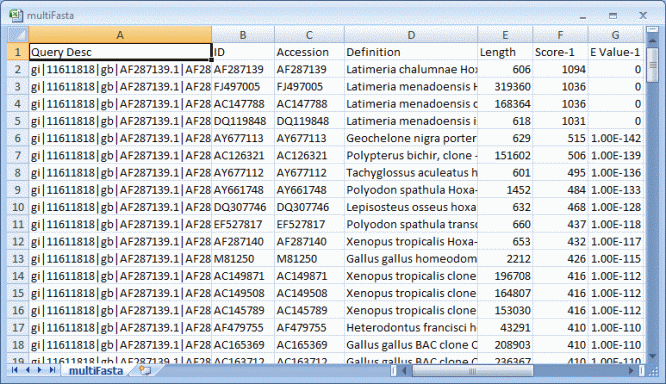
Search results can be exported as an Excel readable CSV file from File > Export
menu. There are "All Results" and "All Best Hits" options.
Export menu is
available when results tab is shown.
Job > Display List menu will display Blast results file in the default
browser.
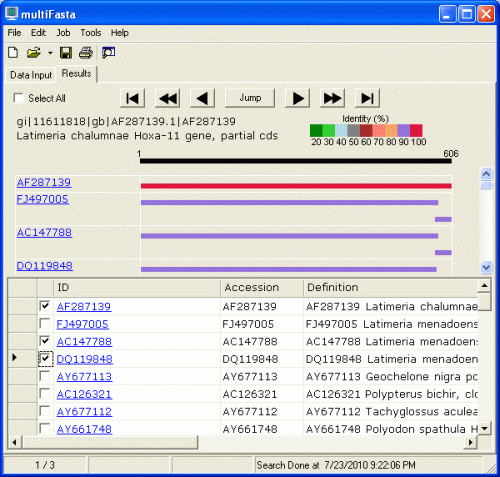
Check check boxes of sequences to be exported. "Select All" will check all
sequences. "Select All" again will uncheck all sequences.
click File > Export FASTA menu. The window below will be shown.
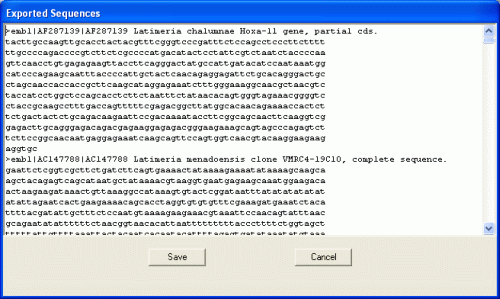
This FASTA data will be saved in the file by clicking "Save". Clicking
"Cancel" will close the window.
More detailed help pages
are available at
http://www.blaststation.com/help/bslocal/en/win/index.html
or Launch
BlastStation-Local and goto Help > Help Menu.
Copyright © 2005 - 2011
TM Software, Inc. All rights
reserved.













 Previous Result
Previous Result Next Result
Next Result Fast Backward
Fast Backward Fast Forward
Fast Forward First Result
First Result Last Result
Last Result



